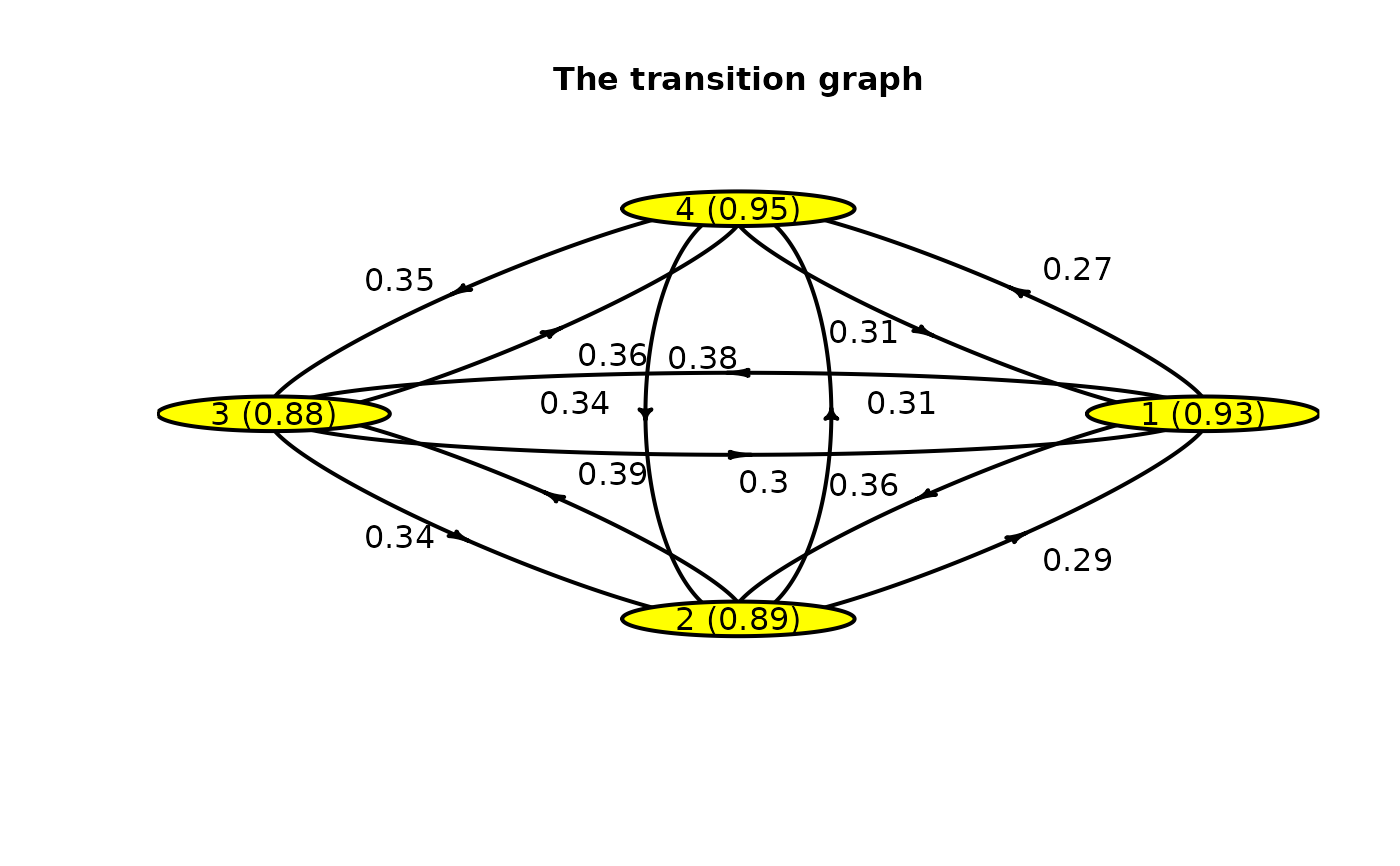
Plot the transition graph between the different states. A node corresponds to a state with the mean time spent in this state. Each arrow represents the probability of transition between states.
# S3 method for class 'Markov'
plot(x, ...)Arguments
- x
output of
estimate_Markovfunction- ...
parameters of
plotmatfunction fromdiagrampackage (see details).
Value
No return value, called for side effects
Details
Some useful extra parameters:
mainmain title.dtextcontrols the position of arrow text relative to arrowhead (default = 0.3).relsizescaling factor for size of the graph (default = 1).box.sizesize of label box, one value or a vector with dimension = number of rows ofx$P.box.cexrelative size of text in boxes, one value or a vector with dimension=number of rows ofx$P.arr.posrelative position of arrowhead on arrow segment/curve.
Examples
# Simulate the Jukes-Cantor model of nucleotide replacement
K <- 4
PJK <- matrix(1 / 3, nrow = K, ncol = K) - diag(rep(1 / 3, K))
lambda_PJK <- c(1, 1, 1, 1)
d_JK <- generate_Markov(n = 100, K = K, P = PJK, lambda = lambda_PJK, Tmax = 10)
# estimation
mark <- estimate_Markov(d_JK)
# transition graph
plot(mark)