Plot categorical functional data
plotData(
data,
group = NULL,
col = NULL,
addId = TRUE,
addBorder = TRUE,
sort = FALSE,
nCol = NULL
)Arguments
- data
data.frame containing
id, id of the trajectory,time, time at which a change occurs andstate, associated state.- group
vector, of the same length as the number individuals of
data, containing group index. Groups are displayed on separate plots. Ifgroup = NA, the corresponding individuals indatais ignored.- col
a vector containing color for each state (can be named)
- addId
If TRUE, add id labels
- addBorder
If TRUE, add black border to each individual
- sort
If TRUE, id are sorted according to the duration in their first state
- nCol
number of columns when
groupis given
Value
a ggplot object that can be modified using ggplot2 package.
On the plot, each row represents an individual over [0:Tmax].
The color at a given time gives the state of the individual.
See also
Other Descriptive statistics:
boxplot.timeSpent(),
compute_duration(),
compute_number_jumps(),
compute_time_spent(),
estimate_pt(),
hist.duration(),
hist.njump(),
plot.pt(),
statetable(),
summary_cfd()
Examples
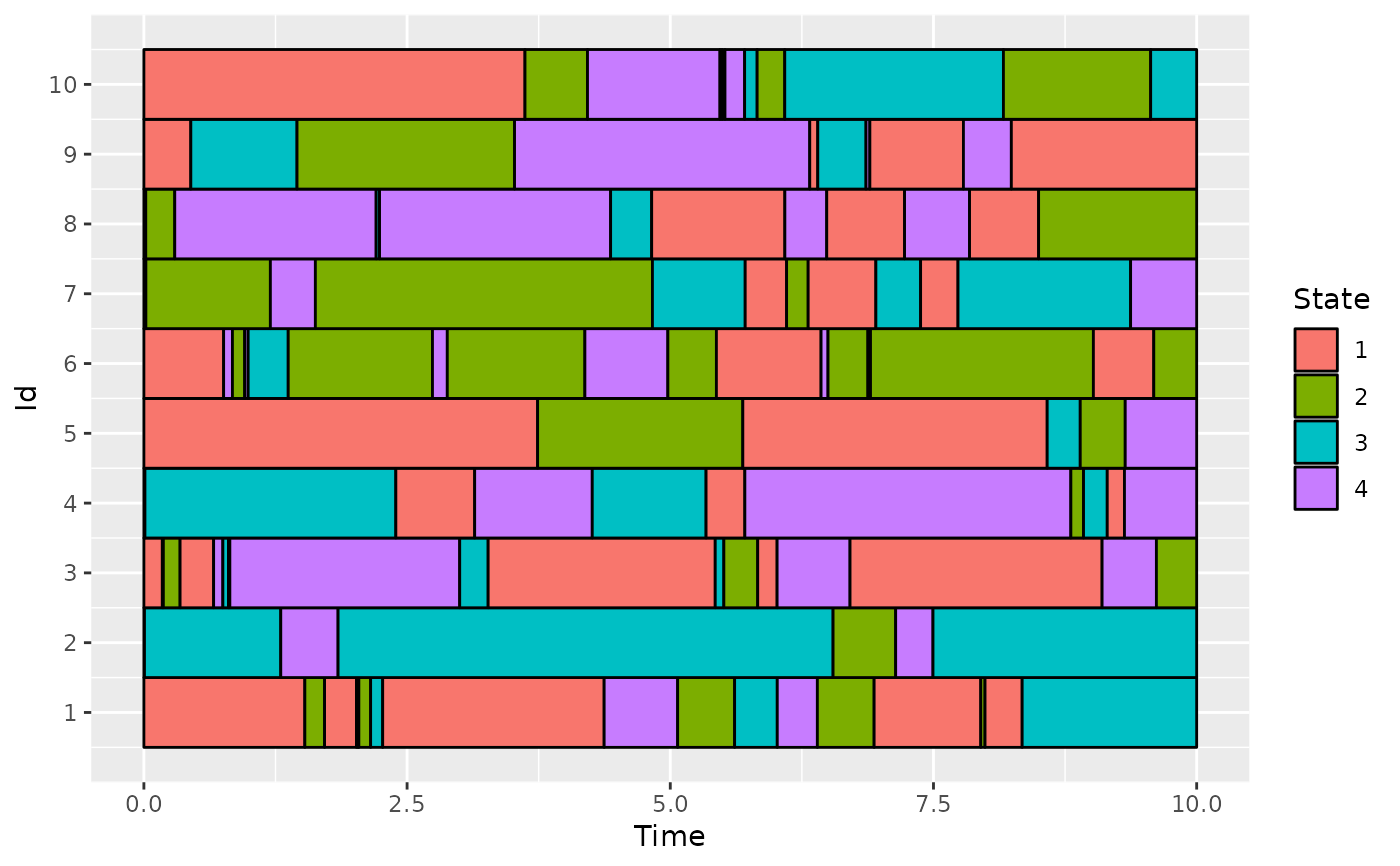
# Simulate the Jukes-Cantor model of nucleotide replacement
K <- 4
PJK <- matrix(1 / 3, nrow = K, ncol = K) - diag(rep(1 / 3, K))
lambda_PJK <- c(1, 1, 1, 1)
d_JK <- generate_Markov(n = 10, K = K, P = PJK, lambda = lambda_PJK, Tmax = 10)
# add a line with time Tmax at the end of each individual
d_JKT <- cut_data(d_JK, Tmax = 10)
plotData(d_JKT)
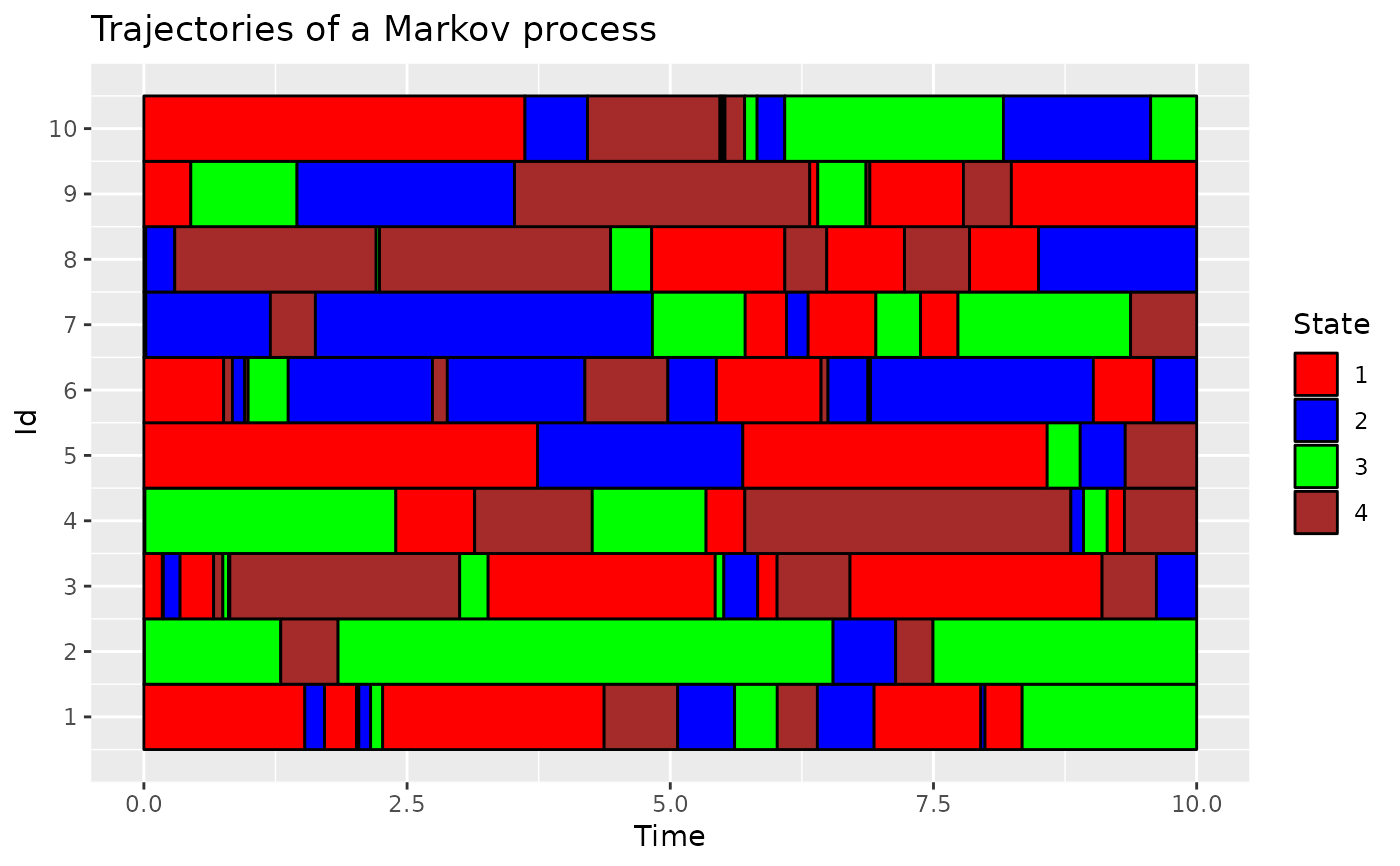
 # modify the plot using ggplot2
library(ggplot2)
plotData(d_JKT, col = c("red", "blue", "green", "brown")) +
labs(title = "Trajectories of a Markov process")
# modify the plot using ggplot2
library(ggplot2)
plotData(d_JKT, col = c("red", "blue", "green", "brown")) +
labs(title = "Trajectories of a Markov process")
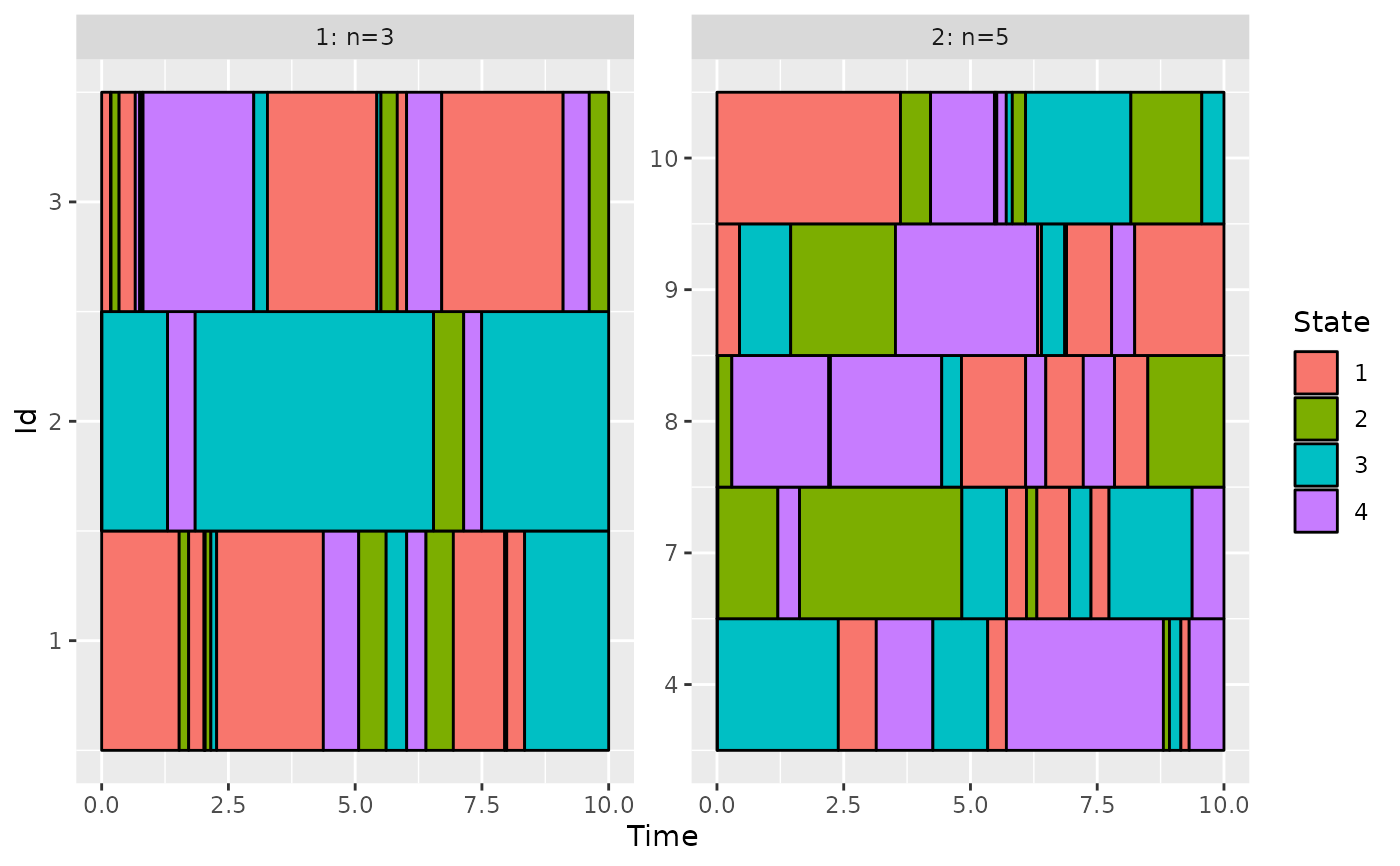
 # use the group variable: create a group with the 3 first variables and one with the others
group <- rep(1:2, c(3, 7))
plotData(d_JKT, group = group)
# use the group variable: create a group with the 3 first variables and one with the others
group <- rep(1:2, c(3, 7))
plotData(d_JKT, group = group)
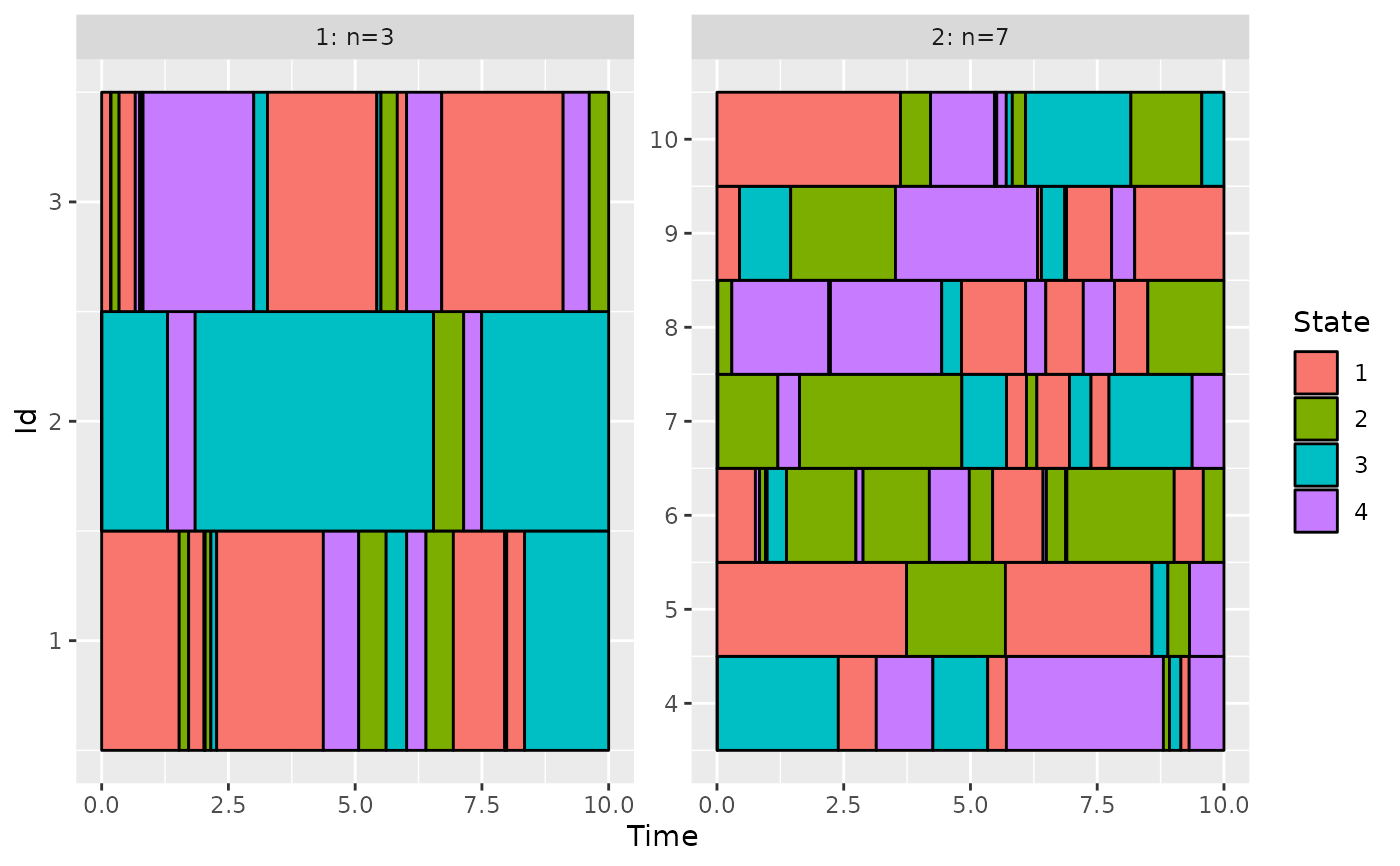 # use the group variable: remove the id number 5 and 6
group[c(5, 6)] <- NA
plotData(d_JKT, group = group)
# use the group variable: remove the id number 5 and 6
group[c(5, 6)] <- NA
plotData(d_JKT, group = group)